Research details
1. Characterization of the azorhizobial chromosome partition proteins : role in regulation of stem-nodule maturation
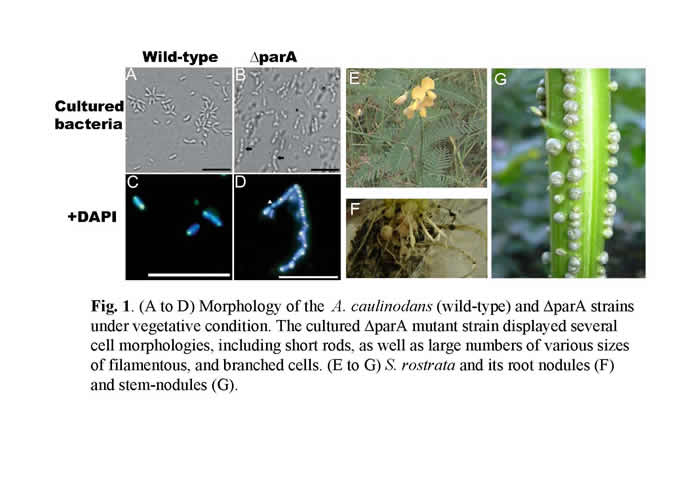
Azorhizobium caulinodans, a Gram-negative a-proteobacterium, possesses the smallest genome size (5.37 Mb) among the known rhizobia (Fig.1). It is the one of few rhizobia that fixes nitrogen ex-planta at a relatively high oxygen concentration. In the symbiotic interaction between A. caulinodans and its host plant, the semi-aquatic tropical legume Sesbania rostrata, nodules are formed not only on the roots but are also found on the stem (Fig.1).
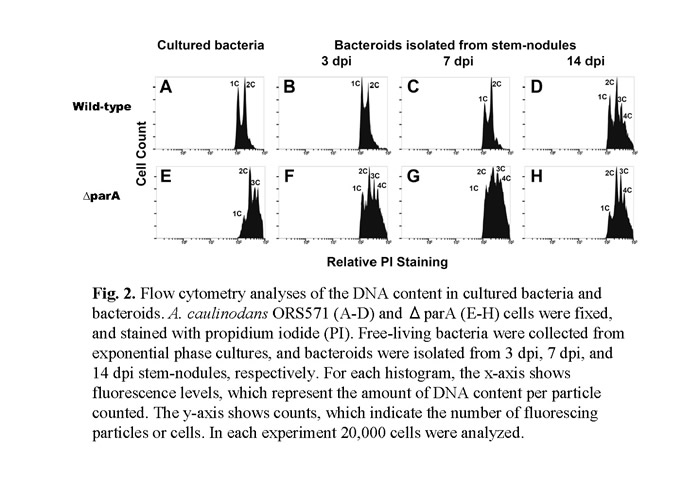
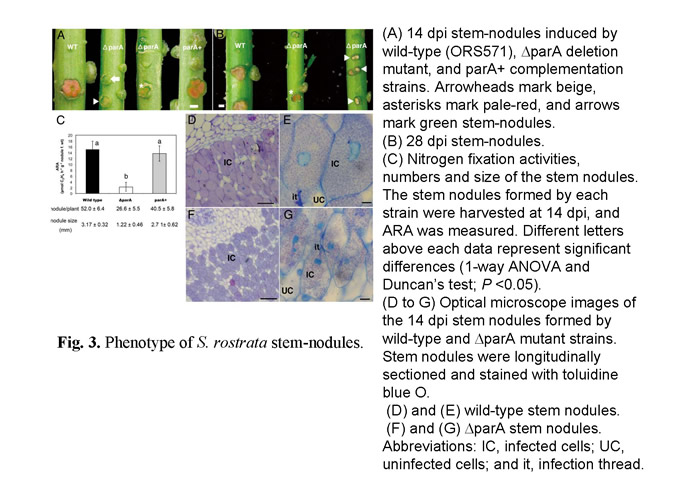
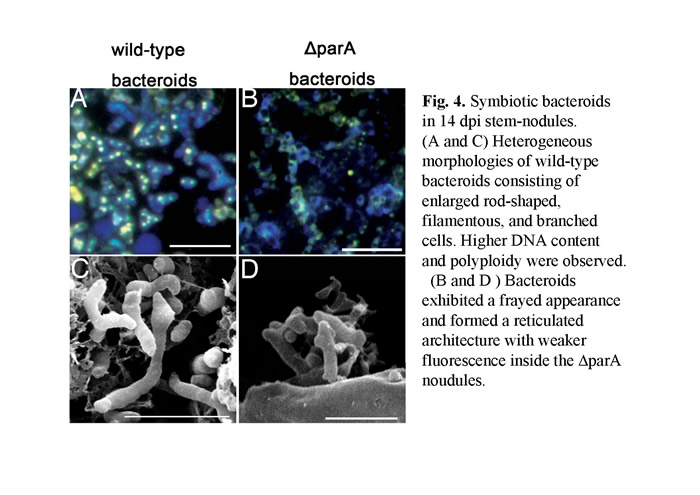
In a previous study (Suzuki et al., 2007), we have identified many novel genes which are involved in the developmental process of nodulation through a large scale in vivo screen for A.caulinodans mutants. We noticed that a null mutant of azorhizobial chromosome partition gene (parA) resulting in notable defects in chromosome partition (Figs.1). Besides, parA defective mutant had already been differentiated untimely before they invading the host plant (Fig. 2). While S. rostrata was inoculated with such mutant strain, immature stem-nodules were generated (Fig.3). The development of the aberrant stem-nodules was halted in diverse maturity stage. A number of bacteroids exhibited a frayed appearance and formed a reticulated architecture with weaker fluorescence inside the parA mutant noudules (Fig.4).
In this project, we have been using a combination of molecular, biochemical, cell biology, genetic and proteomic approaches to elucidate how the azorhizobial chromosome partition system (including ParA and ParB proteins) initiates the bacteroid differentiation process during nodulation. We also expect to identify the specific plant signal molecules from S. rostrata, understand how rhizobia perceive these signals and how they transduce symbiotic signals to downstream components. We hope the findings of this study will serve as a foundation for conferring symbiotic nitrogen fixation to non-legume plants.